Figure 4
Last updated: 2024-12-24
Checks: 5 1
Knit directory: proj_distal/analysis/
This reproducible R Markdown analysis was created with workflowr (version 1.7.1). The Checks tab describes the reproducibility checks that were applied when the results were created. The Past versions tab lists the development history.
Great job! The global environment was empty. Objects defined in the global environment can affect the analysis in your R Markdown file in unknown ways. For reproduciblity it’s best to always run the code in an empty environment.
The command set.seed(12345) was run prior to running the code in the R Markdown file. Setting a seed ensures that any results that rely on randomness, e.g. subsampling or permutations, are reproducible.
Great job! Recording the operating system, R version, and package versions is critical for reproducibility.
Nice! There were no cached chunks for this analysis, so you can be confident that you successfully produced the results during this run.
Great job! Using relative paths to the files within your workflowr project makes it easier to run your code on other machines.
Tracking code development and connecting the code version to the results is critical for reproducibility. To start using Git, open the Terminal and type git init in your project directory.
This project is not being versioned with Git. To obtain the full reproducibility benefits of using workflowr, please see ?wflow_start.
# scRNA-seq
library(Seurat)
packageVersion("Seurat")[1] '4.1.0'# Plotting
library("ggplot2")
library("ggpubr")
library("cowplot")
# Presentation
library("knitr")
# Others
library("readxl")
library("DT")
library("dplyr")
library('stringr')source(here::here("R/00_generalDeps.R"))
source(here::here("R/output.R"))filt_path <- here::here("data/processed/figure4_input_seurat.rds")Introduction
In this document we are going to perform plotting figure4 panelC&D on the high-quality filtered dataset using Seurat.
Loading seurat object processed
if (file.exists(filt_path)) {
patient.integrated <- readRDS(filt_path)
} else {
stop("Figure4 dataset is missing. ",
"Please check Input first.",
call. = FALSE)
}Integration
Integrate data from the six samples, so that cells from the same cell type/subpopulation will cluster together.
visualization
plot metadata
#DimPlot(patient.integrated, reduction = "umap", group.by = "SortPheno") + ggtitle(paste0("SortPheno: ", length(patient.integrated$SortPheno)))
clusterCount = as.data.frame( table( ClusterNumber = patient.integrated[[ "SortPheno" ]]), responseName = "CellCount");
# Create datatable
datatable( clusterCount,
class = "compact",
rownames = FALSE,
colnames = c("SortPheno", "Nb. Cells"),
options = list(dom = "<'row'rt>", # Set elements for CSS formatting ('<Blf><rt><ip>')
autoWidth = FALSE,
columnDefs = list( # Center all columns
list( targets = 0:(ncol(clusterCount)-1),
className = 'dt-center')),
orderClasses = FALSE, # Disable flag for CSS to highlight columns used for ordering (for performance)
paging = FALSE, # Disable pagination (show all)
processing = TRUE,
scrollCollapse = TRUE,
scroller = TRUE, # Only load visible data
scrollX = TRUE,
scrollY = "525px",
stateSave = TRUE));#DimPlot(patient.integrated, reduction = "umap", group.by = "Sample") + ggtitle(paste0("Sample: ", length(patient.integrated$Sample)))
clusterCount = as.data.frame( table( ClusterNumber = patient.integrated[[ "Sample" ]]), responseName = "CellCount");
# Create datatable
datatable( clusterCount,
class = "compact",
rownames = FALSE,
colnames = c("Sample", "Nb. Cells"),
options = list(dom = "<'row'rt>", # Set elements for CSS formatting ('<Blf><rt><ip>')
autoWidth = FALSE,
columnDefs = list( # Center all columns
list( targets = 0:(ncol(clusterCount)-1),
className = 'dt-center')),
orderClasses = FALSE, # Disable flag for CSS to highlight columns used for ordering (for performance)
paging = FALSE, # Disable pagination (show all)
processing = TRUE,
scrollCollapse = TRUE,
scroller = TRUE, # Only load visible data
scrollX = TRUE,
scrollY = "525px",
stateSave = TRUE));Plate4 01-018 issue
Visual plate4 sample 01-018 then filter out
DefaultAssay(patient.integrated) <- 'RNA'
metadata_df = patient.integrated@meta.data
ids = metadata_df[metadata_df$Plate == 'p4' & metadata_df$Sample == '01-018',]$UniqueCellID
metadata_df$p4018 = 'no'
metadata_df[ids, ]$p4018 = 'yes'
patient.integrated$p4018 = metadata_df$p4018
#DimPlot(patient.integrated, reduction = "umap", group.by = "p4018") + ggtitle("Plate 4 018 issue")
Idents(patient.integrated) <- 'p4018'
patient.integrated = subset(patient.integrated, idents = 'yes',invert = TRUE)visualization 2
plot metadata after filter out
#DimPlot(patient.integrated, reduction = "umap", group.by = "SortPheno") + ggtitle(paste0("After filtering Plate 4 018 issue SortPheno: ", length(patient.integrated$SortPheno)))
clusterCount = as.data.frame( table( ClusterNumber = patient.integrated[[ "SortPheno" ]]), responseName = "CellCount");
# Create datatable
datatable( clusterCount,
class = "compact",
rownames = FALSE,
colnames = c("SortPheno", "Nb. Cells"),
options = list(dom = "<'row'rt>", # Set elements for CSS formatting ('<Blf><rt><ip>')
autoWidth = FALSE,
columnDefs = list( # Center all columns
list( targets = 0:(ncol(clusterCount)-1),
className = 'dt-center')),
orderClasses = FALSE, # Disable flag for CSS to highlight columns used for ordering (for performance)
paging = FALSE, # Disable pagination (show all)
processing = TRUE,
scrollCollapse = TRUE,
scroller = TRUE, # Only load visible data
scrollX = TRUE,
scrollY = "525px",
stateSave = TRUE));#DimPlot(patient.integrated, reduction = "umap", group.by = "Sample") + ggtitle(paste0("After filtering Plate 4 018 issue Sample: ", length(patient.integrated$Sample)))
clusterCount = as.data.frame( table( ClusterNumber = patient.integrated[[ "Sample" ]]), responseName = "CellCount");
# Create datatable
datatable( clusterCount,
class = "compact",
rownames = FALSE,
colnames = c("Sample", "Nb. Cells"),
options = list(dom = "<'row'rt>", # Set elements for CSS formatting ('<Blf><rt><ip>')
autoWidth = FALSE,
columnDefs = list( # Center all columns
list( targets = 0:(ncol(clusterCount)-1),
className = 'dt-center')),
orderClasses = FALSE, # Disable flag for CSS to highlight columns used for ordering (for performance)
paging = FALSE, # Disable pagination (show all)
processing = TRUE,
scrollCollapse = TRUE,
scroller = TRUE, # Only load visible data
scrollX = TRUE,
scrollY = "525px",
stateSave = TRUE));Box plots of gene set score
loading markers table
markers table 1 : figure 2, marker table 2 : figure 3
# DEA by Anno_1
dea_f = read.csv(file = here::here("data/processed/figure2_group_gene_markers.csv"), header = TRUE)
Top50_dea_f = dea_f %>% group_by(cluster) %>% top_n(n = 50, wt = avg_log2FC)
selfAg_df <- Top50_dea_f[Top50_dea_f$cluster == "SLA",]
flu_df <- Top50_dea_f[Top50_dea_f$cluster == "FLU",]
#marker2
amr053_df = read_excel(here::here("data/processed/supp_Table7_Rev1.xlsx"), sheet = "Feuil1")
amr053_df = na.omit(amr053_df)Box plots
Gene set score Auto-reactivity & H1N1
for(name in c('selfAg', 'flu')){
df = eval(parse(text = paste0(name, '_df')))
patient.integrated <- AddModuleScore(patient.integrated, features = list(df$gene) ,name = name)
}
metadata_df = patient.integrated@meta.data
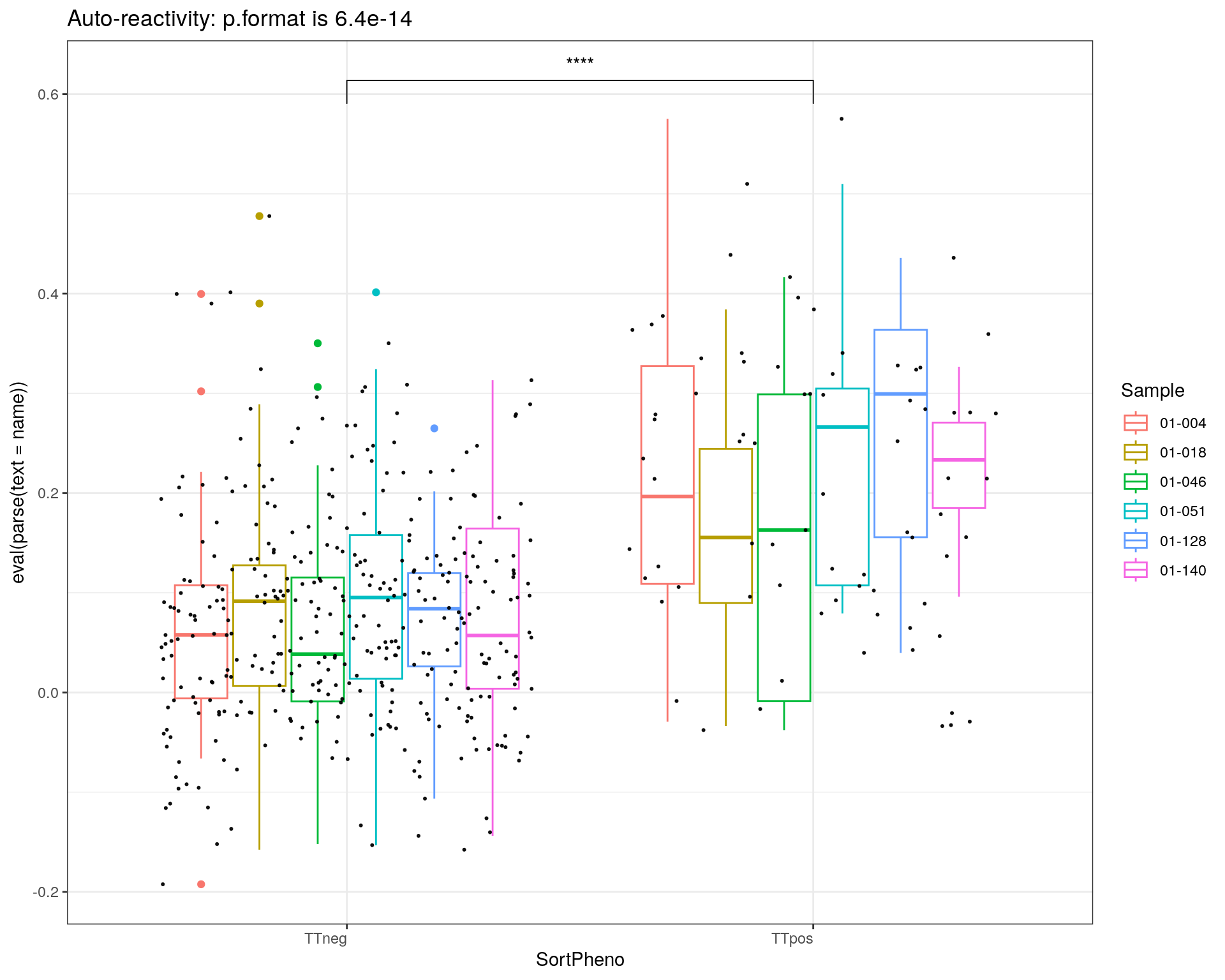
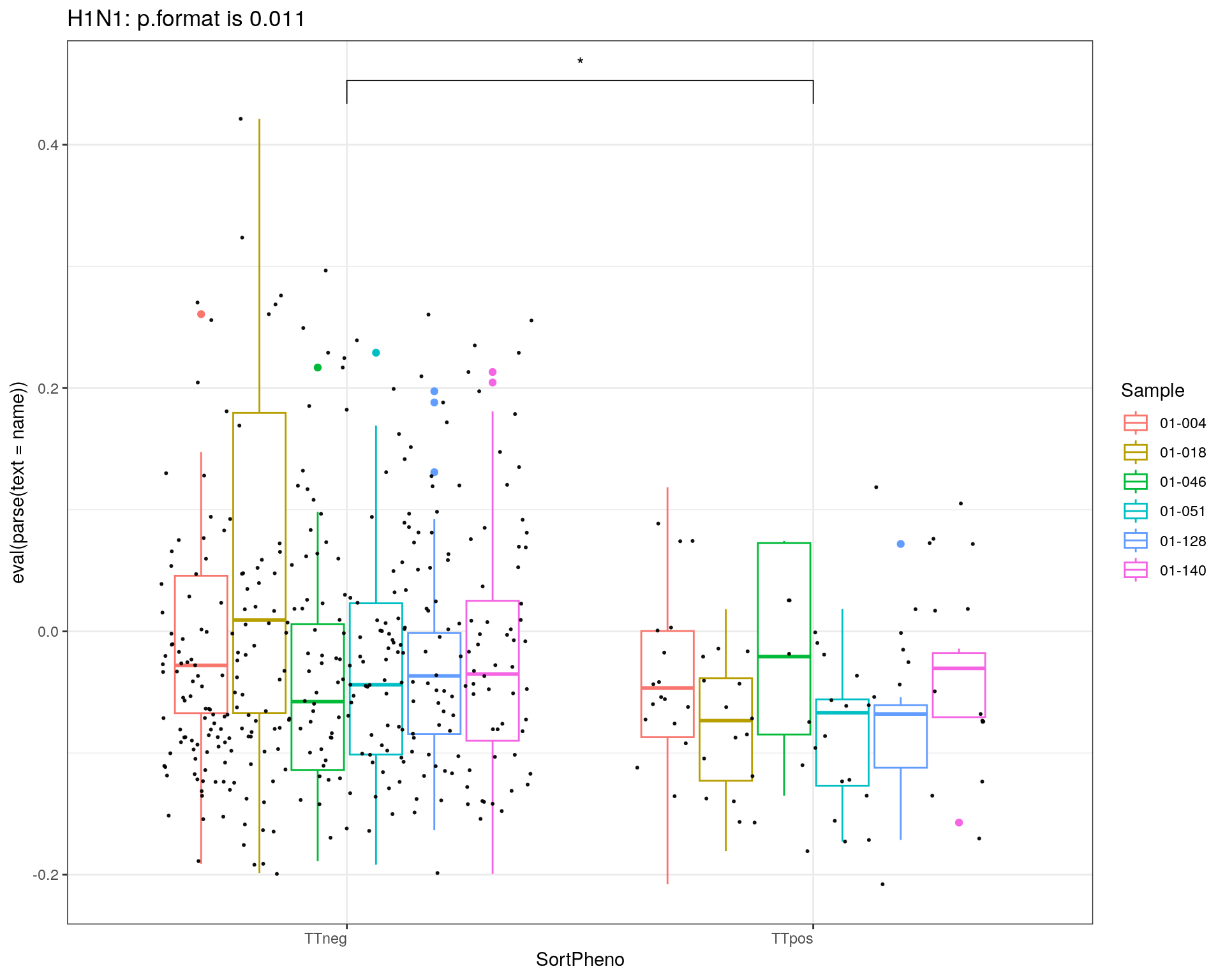
title = c("Auto-reactivity", "H1N1")
i = 1
for(name in c('selfAg1','flu1')){
p<-ggplot(metadata_df, aes(x=SortPheno, y= eval(parse(text = name)), color=Sample)) +
geom_boxplot()+
#scale_color_manual(values=cols) +
geom_jitter(color="black", size=0.4, alpha=0.9) +
theme_bw() +
theme(axis.text.x = element_text(angle = 0, vjust = 1, hjust=1))
my_comparisons <- list(c("TTneg" , "TTpos"))
p <- p + stat_compare_means(comparisons = my_comparisons, label = "p.signif")
if ( i == 1 ){
stat = compare_means(selfAg1 ~ SortPheno, data = metadata_df)
}else{
stat = compare_means(flu1 ~ SortPheno, data = metadata_df)
}
print(p + ggtitle(paste0(title[i],": p.format is ", stat$p.format)))
i = i + 1
}
Cluster markers
for(name in c(1:3,5,6)){
genes = amr053_df[amr053_df$cluster == name,]$gene
if(length(genes) > 50){
genes = genes[1:50]
}
patient.integrated <- AddModuleScore(patient.integrated, features = list(genes) ,name = paste0('cl', name))
}
metadata_df = patient.integrated@meta.data
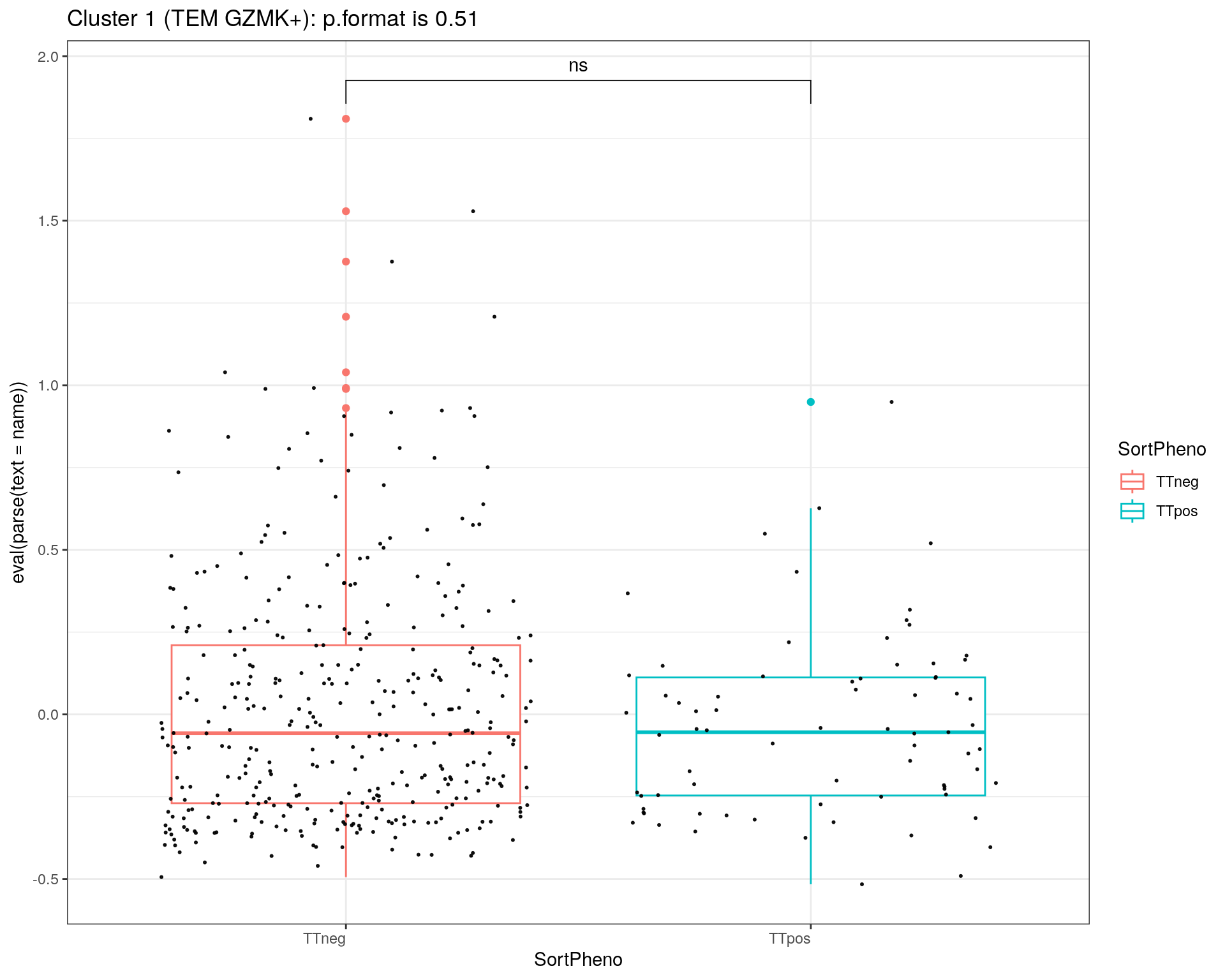
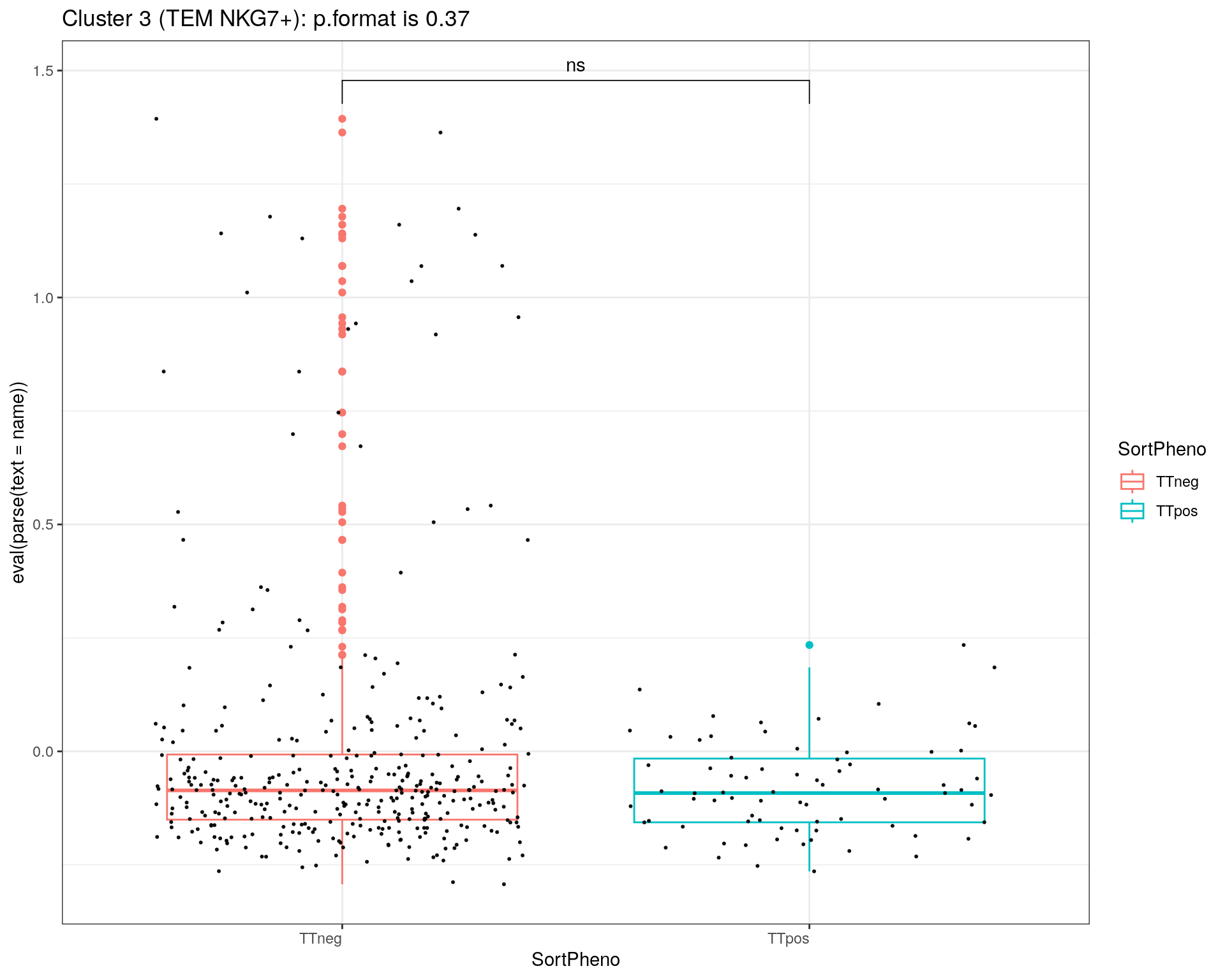
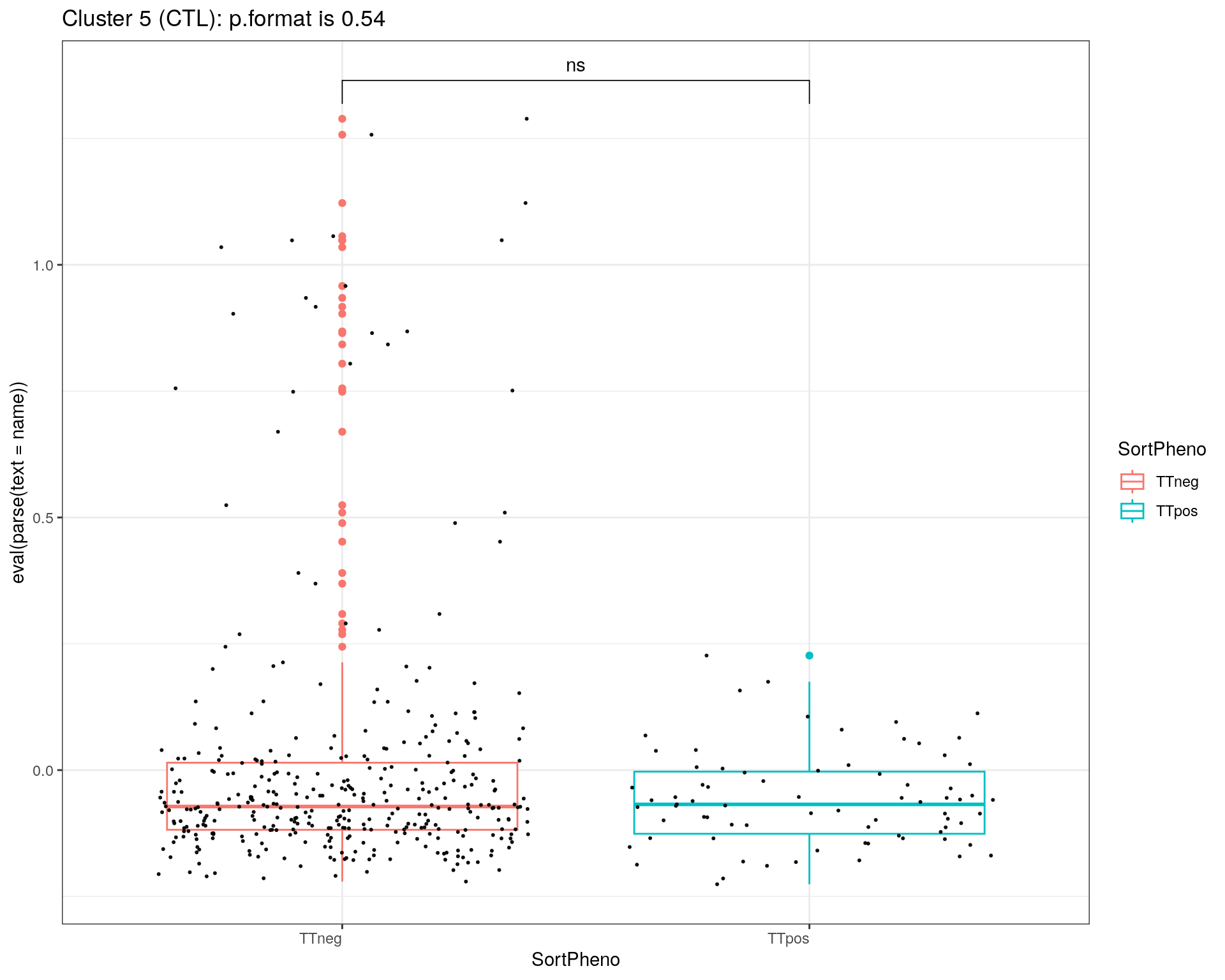
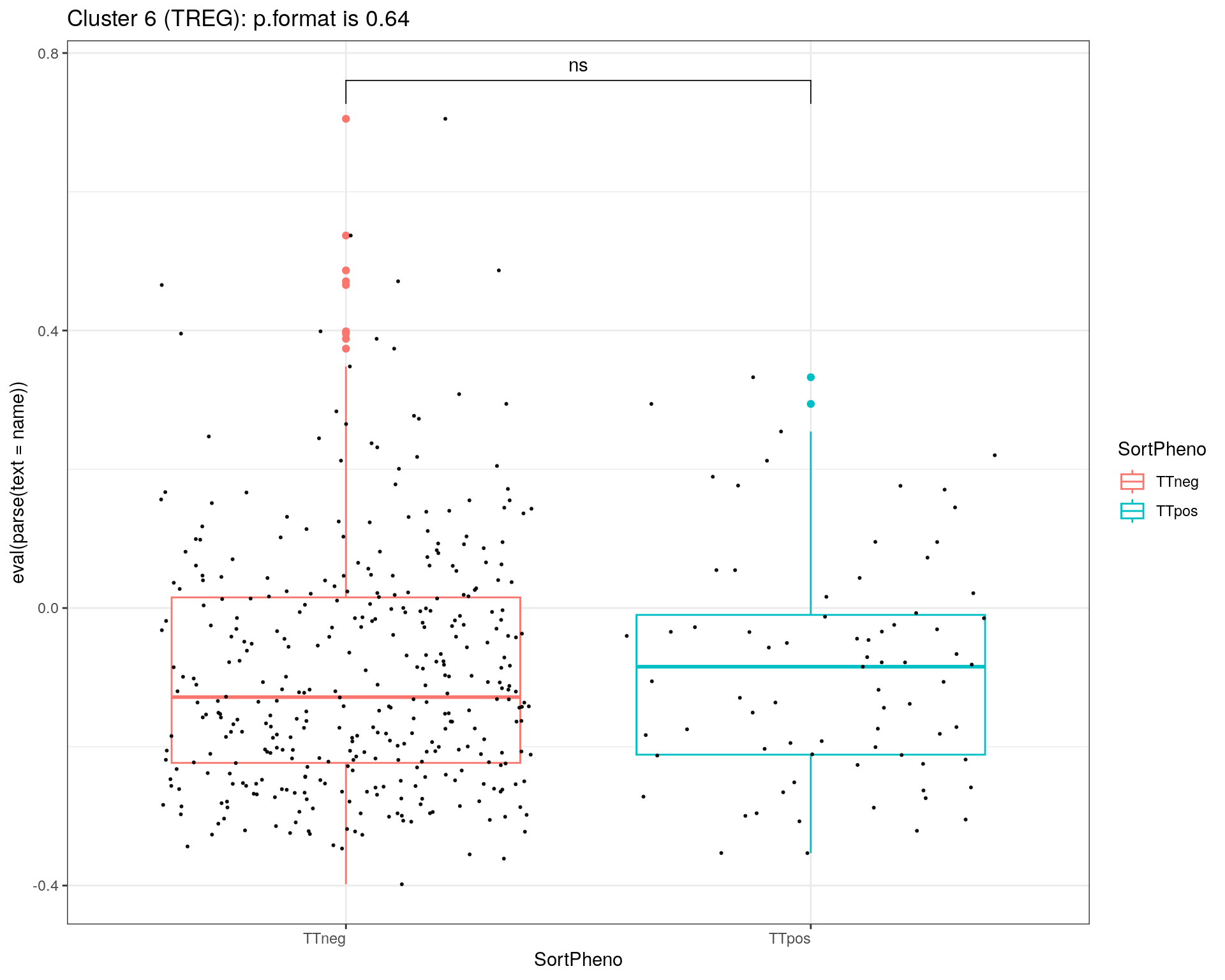
title = c("Cluster 1 (TEM GZMK+)", "Cluster 2 (TCM HLA-DR+)", "Cluster 3 (TEM NKG7+)", "Cluster 5 (CTL)", "Cluster 6 (TREG)")
i = 1
for(name in paste0('cl', c(1:3,5,6), '1')){
p<-ggplot(metadata_df, aes(x=SortPheno, y= eval(parse(text = name)), color=SortPheno)) +
geom_boxplot()+
#scale_color_manual(values=cols) +
geom_jitter(color="black", size=0.4, alpha=0.9) +
theme_bw() +
theme(axis.text.x = element_text(angle = 0, vjust = 1, hjust=1))
my_comparisons <- list(c("TTneg" , "TTpos"))
p <- p + stat_compare_means(comparisons = my_comparisons, label = "p.signif")
if ( i == 1 ){
stat = compare_means(cl11 ~ SortPheno, data = metadata_df)
}else if (i == 2){
stat = compare_means(cl21 ~ SortPheno, data = metadata_df)
}else if (i == 3){
stat = compare_means(cl31 ~ SortPheno, data = metadata_df)
}else if (i == 4){
stat = compare_means(cl51 ~ SortPheno, data = metadata_df)
}else if (i == 5){
stat = compare_means(cl61 ~ SortPheno, data = metadata_df)
}
print(p + ggtitle(paste0(title[i],": p.format is ", stat$p.format)))
i = i + 1
}
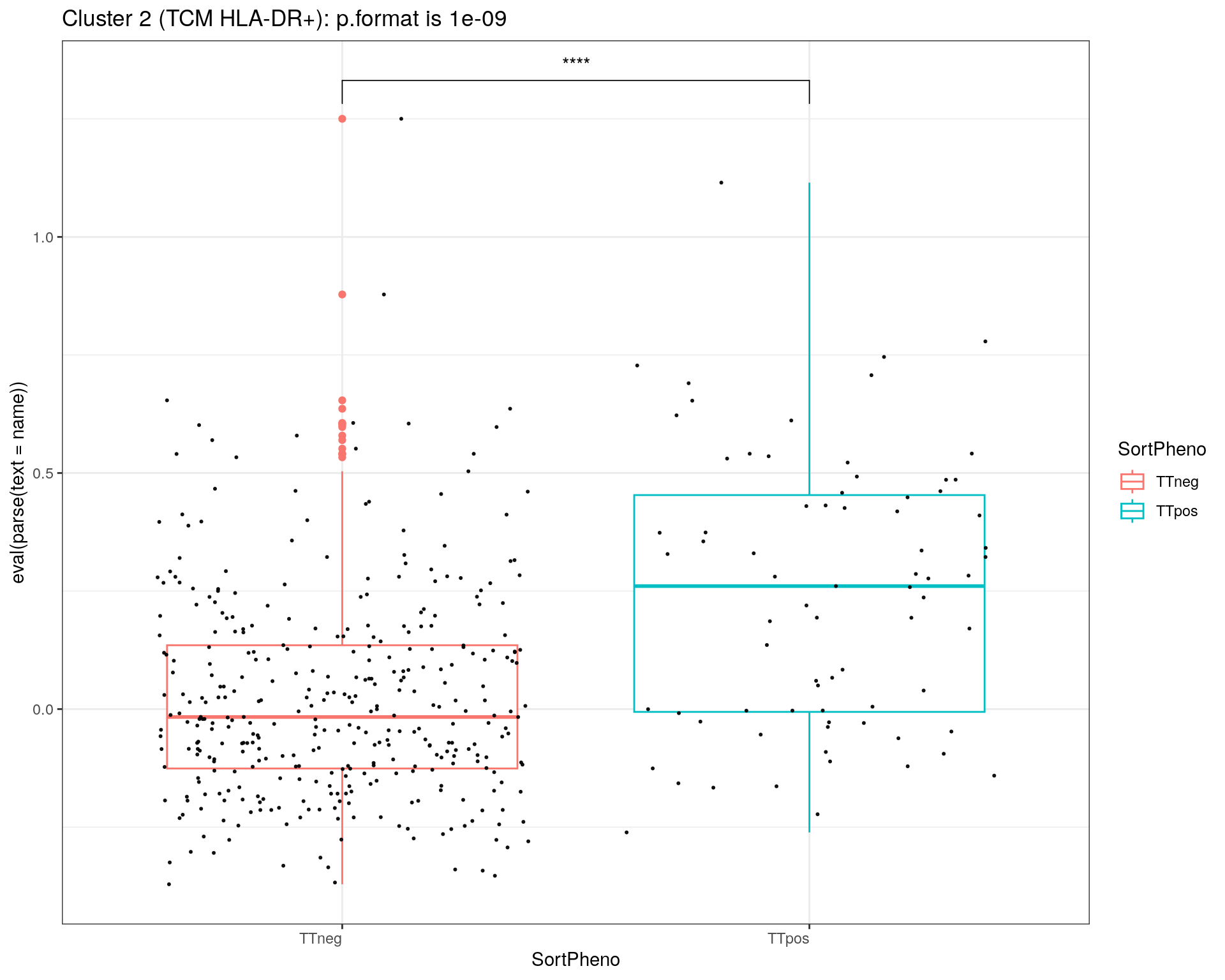
# save scores
df_scores = metadata_df[,c("UniqueCellID", "Sample","selfAg1","flu1","cl11","cl21","cl31","cl51","cl61")]
colnames(df_scores) = c("UniqueCellID", "Sample","Auto-reactivity","H1N1","Cluster 1 (TEM GZMK+)", "Cluster 2 (TCM HLA-DR+)", "Cluster 3 (TEM NKG7+)", "Cluster 5 (CTL)", "Cluster 6 (TREG)")Figure 4
Panel C
my_comparisons <- list(c("TTneg" , "TTpos"))
p1 <-ggplot(metadata_df, aes(x=SortPheno, y= selfAg1, color=Sample)) +
geom_boxplot()+
#scale_color_manual(values=cols) +
geom_jitter(color="black", size=0.4, alpha=0.9) +
theme_bw() +
theme(axis.text.x = element_text(angle = 0, vjust = 1, hjust=1))
p1 <- p1 + stat_compare_means(comparisons = my_comparisons, label = "p.signif")
p1 <- p1 + labs(x = "sorting phenotype", y = "Gene set score", title = "Auto-reactivity")
p2 <-ggplot(metadata_df, aes(x=SortPheno, y= flu1, color=Sample)) +
geom_boxplot()+
#scale_color_manual(values=cols) +
geom_jitter(color="black", size=0.4, alpha=0.9) +
theme_bw() +
theme(axis.text.x = element_text(angle = 0, vjust = 1, hjust=1))
p2 <- p2 + stat_compare_means(comparisons = my_comparisons, label = "p.signif")
p2 <- p2 + labs(x = "sorting phenotype", y = "Gene set score", title = "H1N1")
fig <- plot_grid(p1, p2, nrow = 1, labels = "AUTO")
ggsave(here::here("output", DOCNAME, "figure4-panelC.pdf"), fig,
width = 14, height = 8, scale = 1.2)
ggsave(here::here("output", DOCNAME, "figure4-panelC.png"), fig,
width = 14, height = 8, scale = 1.2)
fig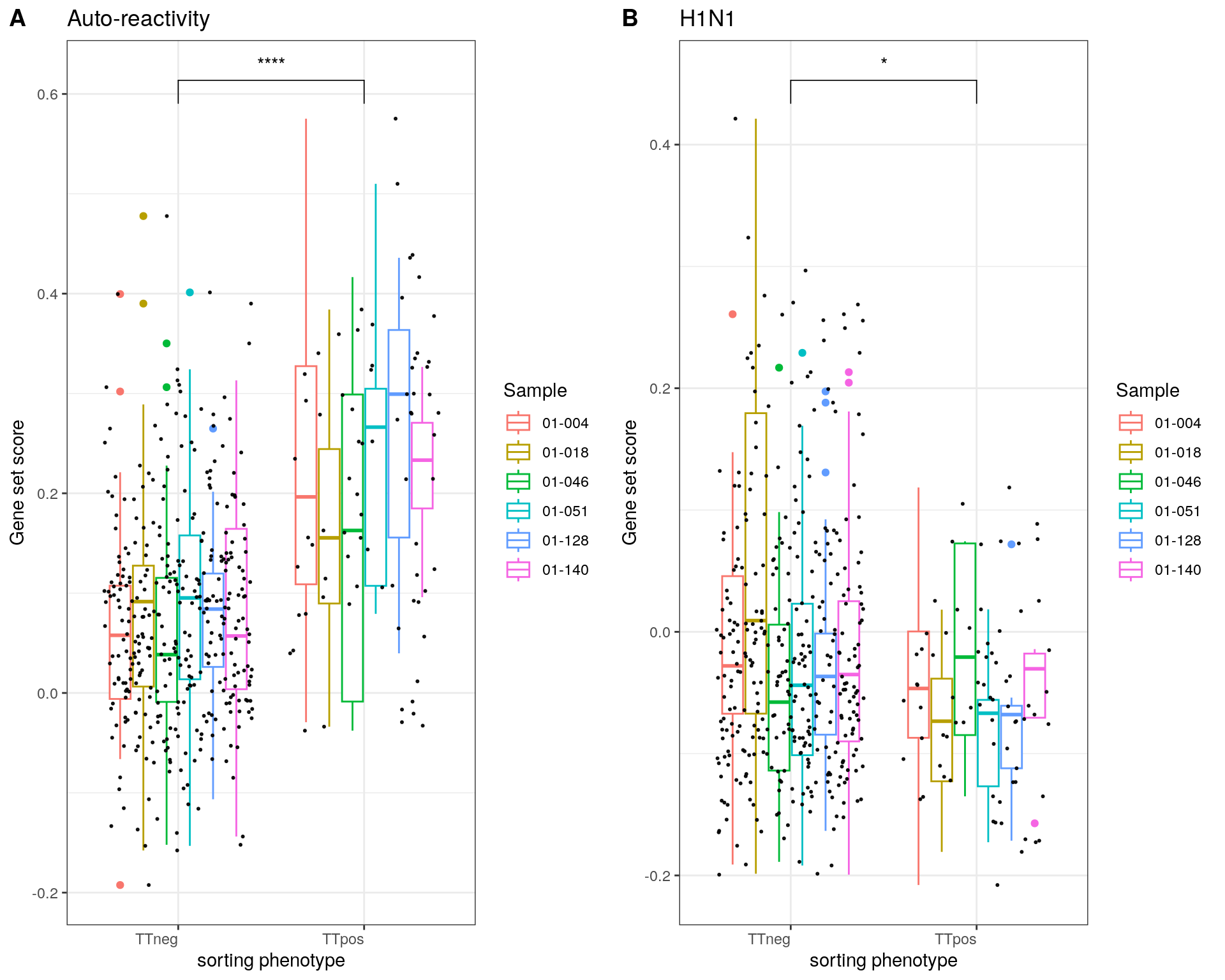
Panel D
p1<-ggplot(metadata_df, aes(x=SortPheno, y= cl11, color=SortPheno)) +
geom_boxplot()+
geom_jitter(color="black", size=0.4, alpha=0.9) +
theme_bw() +
theme(axis.text.x = element_text(angle = 0, vjust = 1, hjust=1))
p1 <- p1 + stat_compare_means(comparisons = my_comparisons, label = "p.signif")
p1 <- p1 + labs(x = "sorting phenotype", y = "Gene set score", title = "Cluster 1 (TEM GZMK+)")
p2<-ggplot(metadata_df, aes(x=SortPheno, y= cl21, color=SortPheno)) +
geom_boxplot()+
geom_jitter(color="black", size=0.4, alpha=0.9) +
theme_bw() +
theme(axis.text.x = element_text(angle = 0, vjust = 1, hjust=1))
p2 <- p2 + stat_compare_means(comparisons = my_comparisons, label = "p.signif")
p2 <- p2 + labs(x = "sorting phenotype", y = "Gene set score", title = "Cluster 2 (TCM HLA-DR+)")
p3<-ggplot(metadata_df, aes(x=SortPheno, y= cl31, color=SortPheno)) +
geom_boxplot()+
geom_jitter(color="black", size=0.4, alpha=0.9) +
theme_bw() +
theme(axis.text.x = element_text(angle = 0, vjust = 1, hjust=1))
p3 <- p3 + stat_compare_means(comparisons = my_comparisons, label = "p.signif")
p3 <- p3 + labs(x = "sorting phenotype", y = "Gene set score", title = "Cluster 3 (TEM NKG7+)")
p4<-ggplot(metadata_df, aes(x=SortPheno, y= cl51, color=SortPheno)) +
geom_boxplot()+
geom_jitter(color="black", size=0.4, alpha=0.9) +
theme_bw() +
theme(axis.text.x = element_text(angle = 0, vjust = 1, hjust=1))
p4 <- p4 + stat_compare_means(comparisons = my_comparisons, label = "p.signif")
p4 <- p4 + labs(x = "sorting phenotype", y = "Gene set score", title = "Cluster 5 (CTL)")
p5<-ggplot(metadata_df, aes(x=SortPheno, y= cl61, color=SortPheno)) +
geom_boxplot()+
geom_jitter(color="black", size=0.4, alpha=0.9) +
theme_bw() +
theme(axis.text.x = element_text(angle = 0, vjust = 1, hjust=1))
p5 <- p5 + stat_compare_means(comparisons = my_comparisons, label = "p.signif")
p5 <- p5 + labs(x = "sorting phenotype", y = "Gene set score", title = "Cluster 6 (Treg)")
fig <- plot_grid(p1, p2, p3, p4, p5, nrow = 2, labels = "AUTO")
ggsave(here::here("output", DOCNAME, "figure4-panelD.pdf"), fig,
width = 14, height = 8, scale = 1.3)
ggsave(here::here("output", DOCNAME, "figure4-panelD.png"), fig,
width = 14, height = 8, scale = 1.3)
fig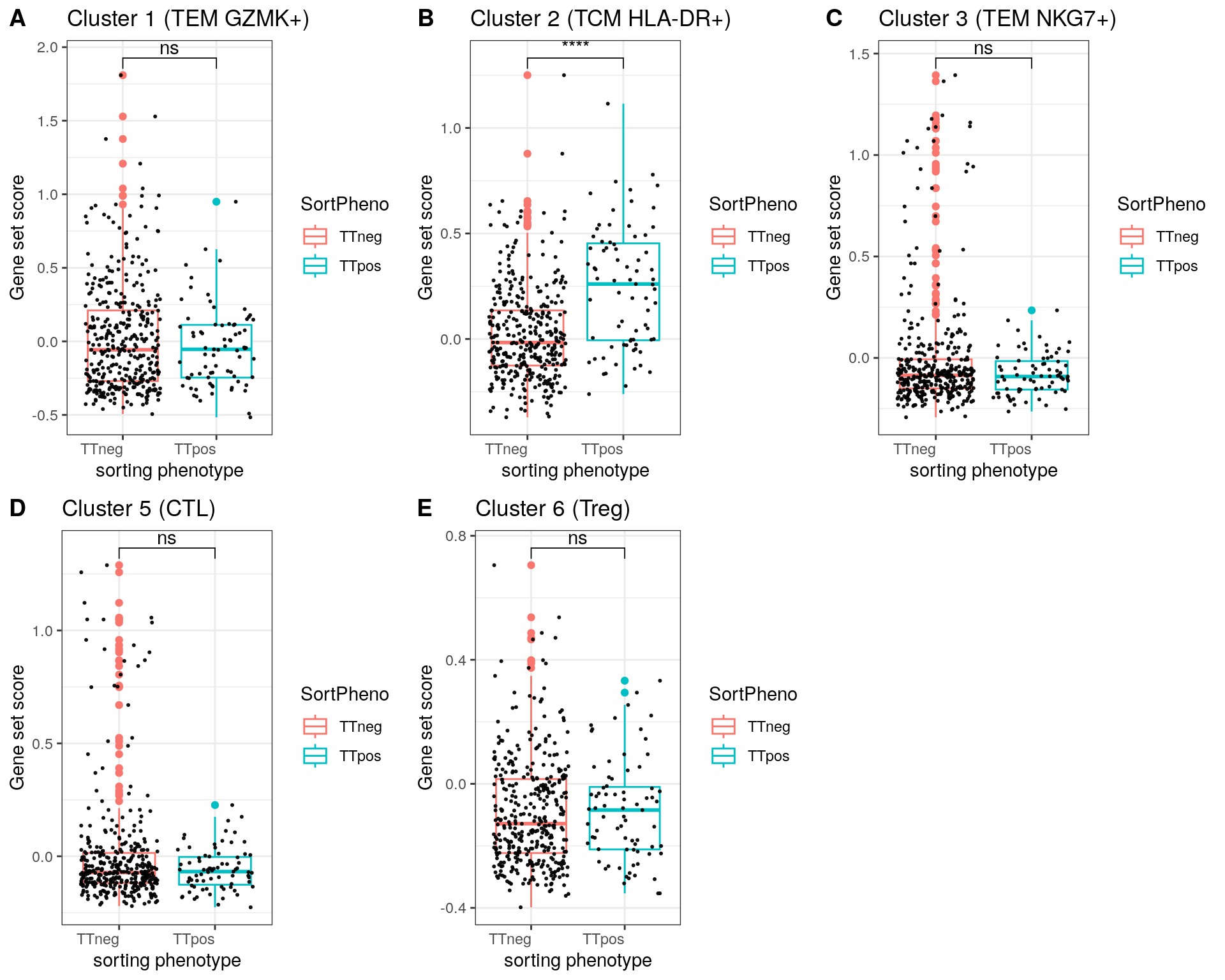
Output files
write.table(df_scores, here::here("output", DOCNAME,"figure4-raw_data.csv"), row.names=FALSE,col.names=TRUE, sep =',')
saveRDS(patient.integrated, here::here("data/processed/figure4_output_seurat.rds"))
save(metadata_df, file = here::here("data/processed/figure4_output_metadata.RData"))Session information
devtools::session_info()─ Session info ───────────────────────────────────────────────────────────────
setting value
version R version 4.1.2 (2021-11-01)
os Ubuntu 20.04.3 LTS
system x86_64, linux-gnu
ui X11
language (EN)
collate en_US.UTF-8
ctype en_US.UTF-8
tz Etc/UTC
date 2024-12-24
pandoc 2.14.0.3 @ /usr/lib/rstudio-server/bin/pandoc/ (via rmarkdown)
─ Packages ───────────────────────────────────────────────────────────────────
package * version date (UTC) lib source
abind 1.4-5 2016-07-21 [1] RSPM (R 4.1.0)
assertthat 0.2.1 2019-03-21 [1] RSPM (R 4.1.0)
backports 1.4.1 2021-12-13 [1] RSPM (R 4.1.0)
brio 1.1.3 2021-11-30 [1] RSPM (R 4.1.0)
broom 0.7.12 2022-01-28 [1] RSPM (R 4.1.0)
bslib 0.3.1 2021-10-06 [1] RSPM (R 4.1.0)
cachem 1.0.6 2021-08-19 [1] RSPM (R 4.1.0)
callr 3.7.0 2021-04-20 [1] RSPM (R 4.1.0)
car 3.0-12 2021-11-06 [1] RSPM (R 4.1.0)
carData 3.0-5 2022-01-06 [1] RSPM (R 4.1.0)
cellranger 1.1.0 2016-07-27 [1] RSPM (R 4.1.0)
cli 3.6.1 2023-03-23 [1] RSPM (R 4.1.0)
cluster 2.1.2 2021-04-17 [2] CRAN (R 4.1.2)
codetools 0.2-18 2020-11-04 [2] CRAN (R 4.1.2)
colorspace 2.0-3 2022-02-21 [1] RSPM (R 4.1.0)
cowplot * 1.1.1 2020-12-30 [1] RSPM (R 4.1.0)
crayon 1.5.0 2022-02-14 [1] RSPM (R 4.1.0)
data.table 1.14.2 2021-09-27 [1] RSPM (R 4.1.0)
DBI 1.1.2 2021-12-20 [1] RSPM (R 4.1.0)
deldir 1.0-6 2021-10-23 [1] RSPM (R 4.1.0)
desc 1.4.1 2022-03-06 [1] RSPM (R 4.1.0)
devtools 2.4.3 2021-11-30 [1] RSPM (R 4.1.0)
digest 0.6.29 2021-12-01 [1] RSPM (R 4.1.0)
dplyr * 1.0.8 2022-02-08 [1] RSPM (R 4.1.0)
DT * 0.22 2022-03-28 [1] RSPM (R 4.1.0)
ellipsis 0.3.2 2021-04-29 [1] RSPM (R 4.1.0)
evaluate 0.15 2022-02-18 [1] RSPM (R 4.1.0)
fansi 1.0.2 2022-01-14 [1] RSPM (R 4.1.0)
fastmap 1.1.0 2021-01-25 [1] RSPM (R 4.1.0)
fitdistrplus 1.1-6 2021-09-28 [1] RSPM (R 4.1.0)
fs 1.5.2 2021-12-08 [1] RSPM (R 4.1.0)
future 1.23.0 2021-10-31 [1] RSPM (R 4.1.0)
future.apply 1.8.1 2021-08-10 [1] RSPM (R 4.1.0)
generics 0.1.2 2022-01-31 [1] RSPM (R 4.1.0)
ggplot2 * 3.4.4 2023-10-12 [1] RSPM (R 4.1.0)
ggpubr * 0.4.0 2020-06-27 [1] RSPM (R 4.1.0)
ggrepel 0.9.1 2021-01-15 [1] RSPM (R 4.1.0)
ggridges 0.5.3 2021-01-08 [1] RSPM (R 4.1.0)
ggsignif 0.6.3 2021-09-09 [1] RSPM (R 4.1.0)
git2r 0.33.0 2023-11-26 [1] RSPM (R 4.1.0)
globals 0.14.0 2020-11-22 [1] RSPM (R 4.1.0)
glue 1.6.2 2022-02-24 [1] RSPM (R 4.1.0)
goftest 1.2-3 2021-10-07 [1] RSPM (R 4.1.0)
gridExtra 2.3 2017-09-09 [1] RSPM (R 4.1.0)
gtable 0.3.0 2019-03-25 [1] RSPM (R 4.1.0)
here 1.0.1 2020-12-13 [1] RSPM (R 4.1.0)
htmltools 0.5.2 2021-08-25 [1] RSPM (R 4.1.0)
htmlwidgets 1.5.4 2021-09-08 [1] RSPM (R 4.1.0)
httpuv 1.6.5 2022-01-05 [1] RSPM (R 4.1.0)
httr 1.4.2 2020-07-20 [1] RSPM (R 4.1.0)
ica 1.0-2 2018-05-24 [1] RSPM (R 4.1.0)
igraph 1.5.1 2023-08-10 [1] RSPM (R 4.1.0)
irlba 2.3.5 2021-12-06 [1] RSPM (R 4.1.0)
jquerylib 0.1.4 2021-04-26 [1] RSPM (R 4.1.0)
jsonlite 1.8.0 2022-02-22 [1] RSPM (R 4.1.0)
KernSmooth 2.23-20 2021-05-03 [2] CRAN (R 4.1.2)
knitr * 1.37 2021-12-16 [1] RSPM (R 4.1.0)
later 1.3.0 2021-08-18 [1] RSPM (R 4.1.0)
lattice 0.20-45 2021-09-22 [2] CRAN (R 4.1.2)
lazyeval 0.2.2 2019-03-15 [1] RSPM (R 4.1.0)
leiden 0.3.9 2021-07-27 [1] RSPM (R 4.1.0)
lifecycle 1.0.3 2022-10-07 [1] RSPM (R 4.1.0)
listenv 0.8.0 2019-12-05 [1] RSPM (R 4.1.0)
lmtest 0.9-39 2021-11-07 [1] RSPM (R 4.1.0)
magrittr 2.0.2 2022-01-26 [1] RSPM (R 4.1.0)
MASS 7.3-54 2021-05-03 [2] CRAN (R 4.1.2)
Matrix 1.3-4 2021-06-01 [2] CRAN (R 4.1.2)
matrixStats 0.61.0 2021-09-17 [1] RSPM (R 4.1.0)
memoise 2.0.1 2021-11-26 [1] RSPM (R 4.1.0)
mgcv 1.8-38 2021-10-06 [2] CRAN (R 4.1.2)
mime 0.12 2021-09-28 [1] RSPM (R 4.1.0)
miniUI 0.1.1.1 2018-05-18 [1] RSPM (R 4.1.0)
munsell 0.5.0 2018-06-12 [1] RSPM (R 4.1.0)
nlme 3.1-153 2021-09-07 [2] CRAN (R 4.1.2)
parallelly 1.30.0 2021-12-17 [1] RSPM (R 4.1.0)
patchwork 1.1.1 2020-12-17 [1] RSPM (R 4.1.0)
pbapply 1.5-0 2021-09-16 [1] RSPM (R 4.1.0)
pillar 1.7.0 2022-02-01 [1] RSPM (R 4.1.0)
pkgbuild 1.3.1 2021-12-20 [1] RSPM (R 4.1.0)
pkgconfig 2.0.3 2019-09-22 [1] RSPM (R 4.1.0)
pkgload 1.2.4 2021-11-30 [1] RSPM (R 4.1.0)
plotly 4.10.0 2021-10-09 [1] RSPM (R 4.1.0)
plyr 1.8.6 2020-03-03 [1] RSPM (R 4.1.0)
png 0.1-7 2013-12-03 [1] RSPM (R 4.1.0)
polyclip 1.10-0 2019-03-14 [1] RSPM (R 4.1.0)
prettyunits 1.1.1 2020-01-24 [1] RSPM (R 4.1.0)
processx 3.5.2 2021-04-30 [1] RSPM (R 4.1.0)
promises 1.2.0.1 2021-02-11 [1] RSPM (R 4.1.0)
ps 1.6.0 2021-02-28 [1] RSPM (R 4.1.0)
purrr 0.3.4 2020-04-17 [1] RSPM (R 4.1.0)
R6 2.5.1 2021-08-19 [1] RSPM (R 4.1.0)
RANN 2.6.1 2019-01-08 [1] RSPM (R 4.1.0)
RColorBrewer 1.1-2 2014-12-07 [1] RSPM (R 4.1.0)
Rcpp 1.0.8 2022-01-13 [1] RSPM (R 4.1.0)
RcppAnnoy 0.0.19 2021-07-30 [1] RSPM (R 4.1.0)
readxl * 1.3.1 2019-03-13 [1] RSPM (R 4.1.0)
remotes 2.4.2 2021-11-30 [1] RSPM (R 4.1.0)
reshape2 1.4.4 2020-04-09 [1] RSPM (R 4.1.0)
reticulate 1.23 2022-01-14 [1] RSPM (R 4.1.0)
rlang 1.1.1 2023-04-28 [1] RSPM (R 4.1.0)
rmarkdown 2.11 2021-09-14 [1] RSPM (R 4.1.0)
ROCR 1.0-11 2020-05-02 [1] RSPM (R 4.1.0)
rpart 4.1-15 2019-04-12 [2] CRAN (R 4.1.2)
rprojroot 2.0.2 2020-11-15 [1] RSPM (R 4.1.0)
rstatix 0.7.0 2021-02-13 [1] RSPM (R 4.1.0)
rstudioapi 0.13 2020-11-12 [1] RSPM (R 4.1.0)
Rtsne 0.15 2018-11-10 [1] RSPM (R 4.1.0)
sass 0.4.0 2021-05-12 [1] RSPM (R 4.1.0)
scales 1.2.1 2022-08-20 [1] RSPM (R 4.1.0)
scattermore 0.7 2020-11-24 [1] RSPM (R 4.1.0)
sctransform 0.3.3 2022-01-13 [1] RSPM (R 4.1.0)
sessioninfo 1.2.2 2021-12-06 [1] RSPM (R 4.1.0)
Seurat * 4.1.0 2022-01-14 [1] RSPM (R 4.1.0)
SeuratObject * 4.0.4 2021-11-23 [1] RSPM (R 4.1.0)
shiny 1.7.1 2021-10-02 [1] RSPM (R 4.1.0)
spatstat.core 2.3-2 2021-11-26 [1] RSPM (R 4.1.0)
spatstat.data 2.1-2 2021-12-17 [1] RSPM (R 4.1.0)
spatstat.geom 2.4-0 2022-03-29 [1] RSPM (R 4.1.0)
spatstat.sparse 2.1-0 2021-12-17 [1] RSPM (R 4.1.0)
spatstat.utils 2.3-0 2021-12-12 [1] RSPM (R 4.1.0)
stringi 1.7.6 2021-11-29 [1] RSPM (R 4.1.0)
stringr * 1.4.0 2019-02-10 [1] RSPM (R 4.1.0)
survival 3.2-13 2021-08-24 [2] CRAN (R 4.1.2)
tensor 1.5 2012-05-05 [1] RSPM (R 4.1.0)
testthat 3.1.2 2022-01-20 [1] RSPM (R 4.1.0)
tibble 3.1.8 2022-07-22 [1] RSPM (R 4.1.0)
tidyr 1.2.0 2022-02-01 [1] RSPM (R 4.1.0)
tidyselect 1.1.2 2022-02-21 [1] RSPM (R 4.1.0)
usethis 2.1.5 2021-12-09 [1] RSPM (R 4.1.0)
utf8 1.2.2 2021-07-24 [1] RSPM (R 4.1.0)
uwot 0.1.11 2021-12-02 [1] RSPM (R 4.1.0)
vctrs 0.6.4 2023-10-12 [1] RSPM (R 4.1.0)
viridisLite 0.4.0 2021-04-13 [1] RSPM (R 4.1.0)
withr 2.5.0 2022-03-03 [1] RSPM (R 4.1.0)
workflowr 1.7.1 2023-08-23 [1] RSPM (R 4.1.0)
xfun 0.30 2022-03-02 [1] RSPM (R 4.1.0)
xtable 1.8-4 2019-04-21 [1] RSPM (R 4.1.0)
yaml 2.3.5 2022-02-21 [1] RSPM (R 4.1.0)
zoo 1.8-9 2021-03-09 [1] RSPM (R 4.1.0)
[1] /usr/local/lib/R/site-library
[2] /usr/local/lib/R/library
──────────────────────────────────────────────────────────────────────────────
sessionInfo()R version 4.1.2 (2021-11-01)
Platform: x86_64-pc-linux-gnu (64-bit)
Running under: Ubuntu 20.04.3 LTS
Matrix products: default
BLAS/LAPACK: /usr/lib/x86_64-linux-gnu/openblas-pthread/libopenblasp-r0.3.8.so
locale:
[1] LC_CTYPE=en_US.UTF-8 LC_NUMERIC=C LC_TIME=en_US.UTF-8 LC_COLLATE=en_US.UTF-8 LC_MONETARY=en_US.UTF-8 LC_MESSAGES=en_US.UTF-8 LC_PAPER=en_US.UTF-8 LC_NAME=C
[9] LC_ADDRESS=C LC_TELEPHONE=C LC_MEASUREMENT=en_US.UTF-8 LC_IDENTIFICATION=C
attached base packages:
[1] stats graphics grDevices utils datasets methods base
other attached packages:
[1] dplyr_1.0.8 DT_0.22 readxl_1.3.1 knitr_1.37 cowplot_1.1.1 ggpubr_0.4.0 ggplot2_3.4.4 SeuratObject_4.0.4 Seurat_4.1.0
loaded via a namespace (and not attached):
[1] backports_1.4.1 workflowr_1.7.1 systemfonts_1.0.4 plyr_1.8.6 igraph_1.5.1 lazyeval_0.2.2 splines_4.1.2 crosstalk_1.2.0 listenv_0.8.0 scattermore_0.7
[11] usethis_2.1.5 digest_0.6.29 htmltools_0.5.2 fansi_1.0.2 magrittr_2.0.2 memoise_2.0.1 tensor_1.5 cluster_2.1.2 ROCR_1.0-11 remotes_2.4.2
[21] globals_0.14.0 matrixStats_0.61.0 spatstat.sparse_2.1-0 prettyunits_1.1.1 colorspace_2.0-3 ggrepel_0.9.1 textshaping_0.3.6 xfun_0.30 callr_3.7.0 crayon_1.5.0
[31] jsonlite_1.8.0 spatstat.data_2.1-2 survival_3.2-13 zoo_1.8-9 glue_1.6.2 polyclip_1.10-0 gtable_0.3.0 leiden_0.3.9 car_3.0-12 pkgbuild_1.3.1
[41] future.apply_1.8.1 abind_1.4-5 scales_1.2.1 DBI_1.1.2 rstatix_0.7.0 miniUI_0.1.1.1 Rcpp_1.0.8 viridisLite_0.4.0 xtable_1.8-4 reticulate_1.23
[51] spatstat.core_2.3-2 htmlwidgets_1.5.4 httr_1.4.2 RColorBrewer_1.1-2 ellipsis_0.3.2 ica_1.0-2 farver_2.1.0 pkgconfig_2.0.3 sass_0.4.0 uwot_0.1.11
[61] deldir_1.0-6 utf8_1.2.2 here_1.0.1 labeling_0.4.2 tidyselect_1.1.2 rlang_1.1.1 reshape2_1.4.4 later_1.3.0 cellranger_1.1.0 munsell_0.5.0
[71] tools_4.1.2 cachem_1.0.6 cli_3.6.1 generics_0.1.2 broom_0.7.12 devtools_2.4.3 ggridges_0.5.3 evaluate_0.15 stringr_1.4.0 fastmap_1.1.0
[81] ragg_1.2.2 yaml_2.3.5 goftest_1.2-3 processx_3.5.2 fs_1.5.2 fitdistrplus_1.1-6 purrr_0.3.4 RANN_2.6.1 pbapply_1.5-0 future_1.23.0
[91] nlme_3.1-153 mime_0.12 brio_1.1.3 compiler_4.1.2 rstudioapi_0.13 plotly_4.10.0 png_0.1-7 ggsignif_0.6.3 testthat_3.1.2 spatstat.utils_2.3-0
[101] tibble_3.1.8 bslib_0.3.1 stringi_1.7.6 highr_0.9 ps_1.6.0 desc_1.4.1 lattice_0.20-45 Matrix_1.3-4 vctrs_0.6.4 pillar_1.7.0
[111] lifecycle_1.0.3 spatstat.geom_2.4-0 lmtest_0.9-39 jquerylib_0.1.4 RcppAnnoy_0.0.19 data.table_1.14.2 irlba_2.3.5 httpuv_1.6.5 patchwork_1.1.1 R6_2.5.1
[121] promises_1.2.0.1 KernSmooth_2.23-20 gridExtra_2.3 parallelly_1.30.0 sessioninfo_1.2.2 codetools_0.2-18 MASS_7.3-54 assertthat_0.2.1 pkgload_1.2.4 rprojroot_2.0.2
[131] withr_2.5.0 sctransform_0.3.3 mgcv_1.8-38 parallel_4.1.2 grid_4.1.2 rpart_4.1-15 tidyr_1.2.0 rmarkdown_2.11 carData_3.0-5 Rtsne_0.15
[141] git2r_0.33.0 shiny_1.7.1